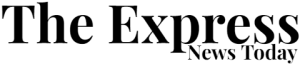Set-up of Rb-Tn-seq experiments
We constructed Rb-Tn-seq libraries to study Salmonella stress response37 in four phylogenetically distinct and genetically tractable serovars. These included two generalist isolates (S. Typhimurium ST4/74 and D23850) and two human-restricted isolates (S. Typhi Ty2 and S. Paratyphi A 9150). On average, each library contained 166,905 unique genome-wide transposon insertion sites which integrated every 27.7 bp. The median insertions per gene ranged from 12 to 44 across these serovars, with central Tn insertions in ~90–92% of coding genes. Barcoded transposons showed even distribution across chromosomes and plasmids, ensuring high genome coverage with minimal strand or coverage bias (Fig. 1b, Supplementary Fig. 1a–e and Supplementary Table 4). The insertion index of each gene was calculated to identify 427 to 476 putative essential genes for each serovar (Supplementary Data 1 and Supplementary Fig. 2a–d), several of which were serovar-specific, including igaA in Typhimurium ST4/74 and D23580, rpoE in S. Typhi Ty2 and various iron homeostasis genes in Paratyphi A (Supplementary Fig. 2e and Supplementary Note 1)38.
We conducted fitness assays on each library, evaluating their response to (1) extracellular stresses encountered in the intestinal tract and/or systemic tissues, (2) intracellular stresses within host cells, including macrophages and (3) exposure to a diverse suite of antibiotics (Fig. 1c and Supplementary Table 5). Stressor concentrations were optimized to achieve ~30–50% growth reduction38 (Fig. 1c and Supplementary Fig. 3a–d). Each experiment included biological duplicates, which were tightly correlated and passed published quality-control metrics36 (Supplementary Fig. 3e–h and Supplementary Data 2).
We used a moderated t-like statistic with |t| > 4 to identify significant fitness events36, leading to the identification of hundreds of genes with significant fitness effects for each serovar across our conditions (Fig. 1d and Supplementary Data 3 and 4). We then used agglomerative clustering to generate heatmaps with all genes with |t| > 4 in at least 1 condition, retaining 678 to 781 genes for each serovar (Supplementary Figs. 4–7). These clustered heatmaps revealed patterns linking similar conditions, including grouping intracellular stresses (for example, InSPI2, InSPI2 Mg, H2O2, NO, bleach), extracellular stresses (for example bile, heat stress, anaerobiosis, gut microbiota media (GMM)) and another grouping linking various antibiotics (for example, ciprofloxacin, azithromycin, rifampicin).
We identified many functionally related gene clusters involved in stress response (Supplementary Data 5). For instance, acrAB/tolC Tn insertions displayed significant fitness defects during bile stress for typhoidal Salmonella (Fig. 1e)39,40. LPS modification arn operon and pmrAB mutations exhibited reduced fitness under polymyxin B in Typhimurium D23580 (Fig. 1e and Supplementary Fig. 8a)41,42,43,44. Mutations in DNA repair genes (for example recDGNQX) led to decreased D23580 fitness with ciprofloxacin—an antibiotic inducing DNA damage (Supplementary Fig. 8b)45,46. Tn insertions in iron homeostasis genes (for example, entDEF, exbD, tonB) caused decreased fitness under iron restriction in Paratyphi A 9150 (Supplementary Fig. 8c). Tn insertions in molybdenum (Mo) metabolism genes (for example moeA, moaA, mog, mobA) showed fitness defects in GMM in several serovars (Supplementary Fig. 8d)47,48. LPS-synthesizing gene mutations (for example, rfaL, rfbBD, wzyO4, waaK) increased sensitivity to both intracellular and extracellular stresses, underscoring the broad role of LPS during bacterial stress response49,50 (Supplementary Fig. 8e). Intriguingly, mutations in barA and sirA exhibited increased fitness under multiple stresses (Supplementary Fig. 8f), possibly explaining their frequent occurrence in chronically infected Salmonella patients51.
Several SPI-encoded genes displayed significant phenotypes (Supplementary Data 6)4. For instance, mutations in hilD—an SPI-1 encoded transcription factor—increased fitness during bile stress in S. Typhi Ty2 and heat shock in S. Typhimurium D23580 (Supplementary Data 6), aligning with a study proposing a role for HilD role in enhancing membrane permeability52. SPI-3-encoded magnesium importers mgtB and mgtC mutations decreased S. Paratyphi A 9150 survival in macrophage-mimicking media InSPI2 Mg (Supplementary Data 6). Mutations in SPI-7-encoded Vi capsule genes in Typhi increased fitness under protamine stress—a positively charged antimicrobial peptide (Supplementary Data 6). Despite Typhi and Paratyphi A encoding unique genes not found in Typhimurium genomes, only a small proportion of these unique genes exhibited significant fitness effects (3.9% in Typhi, 3.2% in Paratyphi A) (Supplementary Data 7). In contrast, a higher proportion of shared orthologs had significant phenotypes in Typhi (18.5%) and Paratyphi A (16.5%) (Supplementary Table 6). To highlight specific processes involved in Salmonella stress response, we sorted significant fitness events by annotated gene ontology (GO) terms and BioCyc-derived functional annotations, grouped into various functional classes (Fig. 1f). Dozens of fitness events involved uncharacterized genes, suggesting that our Rb-Tn-seq dataset holds rich uncharacterized biology (Fig. 1f).
Systems biology approach to analyze fitness profiles
To systematically analyze the thousands of fitness effects captured through Rb-Tn-seq, we employed cofitness network analysis and spatial analysis of functional enrichment (SAFE) to overlay functional data onto network maps38, which has been performed previously in Saccharomyces cerevisiae53 and Streptococcus pneumoniae38. Briefly, we constructed correlation matrices reflecting the log2 fitness changes for each gene across all conditions. These matrices were transformed into cofitness interaction networks, where nodes represented genes and edges indicated correlation values, using a Pearson’s correlation of R > 0.75 to identify closely related fitness profiles (Fig. 2a and Supplementary Data 8). Stability testing38 indicated high significance across our networks (Supplementary Data 8).
a, Left, cofitness network analysis of all genes in S. Typhimurium ST4/74 with r > 0.75; all blue nodes are genes and all gray lines connect pairs of cofit (r > 0.75) genes. Right, SAFE highlights regions of the network that are enriched in certain functional terms. Each colored area represents a different functionally enriched area on the network. b, Examples of subclusters of genes that are identified through SAFE. c,d, Left, cofitness network analysis of all genes in S. Typhi Ty2 (c) and S. Paratyphi A 9150 (d) with r > 0.75; all blue nodes are genes and all gray lines are between pairs of cofit genes. Middle, filtered network that only includes genes with fitness changes that are (1) significant in S. Typhi/S. Paratyphi A but not in S. Typhimurium and (2) > 2-FC in S. Typhi/S. Paratyphi A compared with S. Typhimurium. Right, SAFE highlights regions of the network that are enriched in certain functional terms on these filtered maps. e, An LPS modification cluster including rfbE (purple) is identified through SAFE on the S. Typhi filtered map. f, Heatmap showing the fitness values of Tn insertions in rfbE across 24 plate-based stresses for S. Typhi Ty2. Color gradient is derived from the log2(FC) from each condition in the Rb-Tn-seq experiments. g, Growth curves of Ty2WT (black) and Ty2ΔrfbE (blue) when exposed to 4% bile, with reads taken at OD600 once every 10 min. h, Growth curves of Ty2WT (black) and Ty2ΔrfbE (blue) when exposed to 2.3 µg ml−1 protamine, with reads taken at OD600 once every 10 min. For growth curve experiments (g,h), each point and error bar indicates the mean ± s.e.m. of OD600, derived from n = 4 (g) and n = 3 (h) biologically independent experiments.
SAFE was then applied to annotate each node based on BioCyc classifications and GO terms (Supplementary Data 9)54, through which we identified local network neighborhoods enriched for specific functional classes38,55 (Fig. 2a and Extended Data Fig. 1a–d). To validate this analysis, we searched SAFE outputs for gene networks expected to cluster together based on related functionality, finding clusters for the NADH quinone oxidoreductase complex, molybdenum metabolism genes and genes associated with lipid trafficking to the outer membrane (Fig. 2b). Intriguingly, ~15–20% submodules contain at least one hypothetical gene, including RS16480/STM_3341 in ST4/74, which is correlated strongly with cpxR, and RS03310/t0654 in Typhi Ty2, which is correlated strongly with 24 amino acid metabolism genes (Supplementary Data 10 and Supplementary Note 2)56.
We used the cofitness network pipeline to pinpoint typhoid-specific fitness changes. To this end, we applied an additional filtering step where, for each condition, we only retained genes that (1) had a significant fitness change (|t| > 4) in Typhi Ty2 or Paratyphi A 9150 but not in Typhimurium ST4/74 and (2) had a fold change (FC) that was at least twofold greater in Typhi or Paratyphi compared with Typhimurium (Fig. 2c,d and Supplementary Data 11–12). These filters removed ~75% and 65% of the nodes and connections within each cofitness network for Typhi and Paratyphi, respectively, but still retained thousands of serovar-specific correlations. We then applied SAFE to these filtered networks and identified gene clusters with serovar-specific fitness changes (Fig. 2c,d), including those involved in LPS modification, amino acid metabolism and metal homeostasis. In contrast, only ~3% of the network was retained in D23580 when doing this same analysis, indicating similar fitness profiles between these Typhimurium isolates (Extended Data Fig. 1e and Supplementary Data 13).
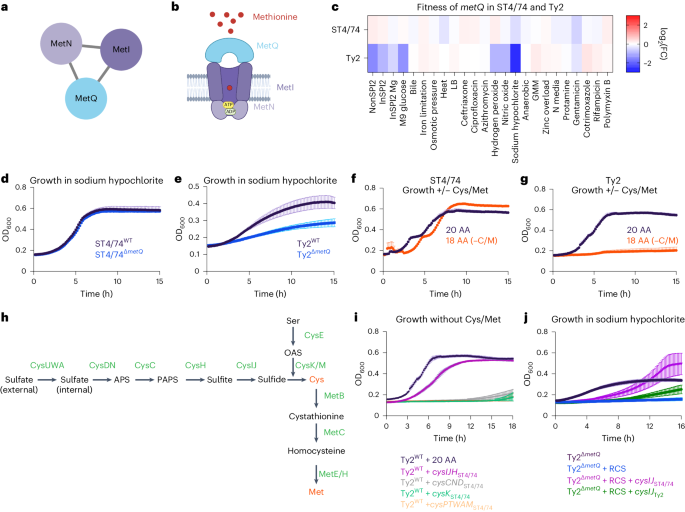
We then examined several gene networks with serovar-specific changes in typhoidal Salmonella, focusing on three categories for further mechanistic investigation. The first includes clusters with genes unique to typhoidal Salmonella, including rfbE (Fig. 2). The second comprises gene clusters where all genes are shared between nontyphoidal and typhoidal Salmonella but exhibit different phenotypes in distinct isolates (for example, metIQN in Fig. 3). The third involves gene clusters containing at least one uncharacterized gene in Salmonella (for example, ybdZ in Fig. 4).
a, metIQN cluster, derived from the S. Typhi Ty2 filtered network. b, Schematic of MetIQN. c, Heatmap showing fitness of metQ Tn insertions across 24 conditions for S. Typhimurium ST4/74 and S. Typhi Ty2. Color gradient is derived from the log2(FC) from Rb-Tn-seq. d, Growth of ST4/74WT (black) and ST4/74ΔmetQ (blue) exposed to 12.5 µg ml−1 sodium hypochlorite, derived from n = 3 biologically independent experiments. e, Growth of Ty2WT (black) and Ty2ΔmetQ (blue) exposed to 12.5 µg ml−1 sodium hypochlorite, derived from n = 3 biologically independent experiments. f,g, Growth of ST4/74WT (f) and Ty2WT(g) when grown in defined minimal medium with a mix of all 20 amino acids added (20 AA; purple), or with 18 amino acids added, without Cys/Met (18AA (−C/M); orange), derived from n = 3 biologically independent experiments. h, Schematic of endogenous synthesis pathway for Cys/Met. Intermediate substrates are shown in black and enzymes are shown in green. Cys/Met are shown in red. i, Growth of WT and complemented strains in which different operons in the endogenous Cys/Met pathway from S. Typhimurium ST4/74 are expressed in S. Typhi Ty2. As a control, Ty2WT was grown in minimal medium supplemented with 20 AA and is shown in black (n = 3 biologically independent experiments). All complementation growth curves were run in minimal medium supplemented with 18AA, but no Cys/Met (−C/M), and are derived from n = 4 biologically independent experiments; magenta, CysIJHST4/74; gray, CysCNDST4/74; green, CysKST4/74; orange, CysAWUMST4/74. j, Growth curves of WT, mutant and complementation strains when exposed to a lethal dose of 25 µg ml−1 sodium hypochlorite. Black, Ty2ΔmetQ growth in the absence of RCS; blue, Ty2ΔmetQ growth with RCS; purple, growth of Ty2ΔmetQ + cysIJST4/74 in the presence of RCS; green, growth of Ty2ΔmetQ + cysIJTy2. All curves are derived from n = 5 biologically independent experiments. For all growth curves (d–g,i,j), each point and error bar indicates the mean ± s.e.m. of OD600 with reads taken at OD600 once every 10 min. APS, adenosine 5′-phosphosulfate; OAS, O-acetyl-Ser; PAPS, 3′-phosphoadenosine 5′-phosphosulfate.
a, Cluster of genes involved in metal homeostasis, identified through SAFE on the S. Paratyphi A 9150 filtered network. b, Transcript levels of ybdZ measured by qRT-PCR and normalized to a control gene (rpoD). Bars indicate mean ± s.e.m., with individual measurements shown (black dots). Each bar is derived from n = 3 biologically independent experiments. c, Heatmap showing the fitness values of Tn insertions in ybdZ across 24 stress conditions for S. Typhimurium ST4/74 and S. Paratyphi A 9150. Color gradient is derived from the log2(FC) from each condition in the Rb-Tn-seq experiments. d, Growth curves of ST4/74WT (black) and ST4/74ΔybdZ (blue) when exposed to 100 µM 2,2′-dipyridyl, +/− addition of exogenous 1 mM FeCl3 (orange), with reads taken at OD600 once every 10 min, derived from n = 3 biologically independent experiments. e, Growth curves of Para9150WT (black) and Para9150ΔybdZ (blue) when exposed to 100 µM 2,2′-dipyridyl, +/− addition of exogenous 1 mM FeCl3 (orange), with reads taken at OD600 once every 10 min, derived from n = 3 biologically independent experiments. f, Growth curves of Para9150ΔybdZ and complementation strains in which functional versions of different iron-related pseudogenes from S. Typhimurium ST4/74 are expressed in Para9150ΔybdZ under iron restriction, with reads taken at OD600 once every 10 min; SUFST4/74 is shown in red, FhuAST4/74 is shown in green, and FhuEST4/74 is shown in purple. For controls, the growth of Para9150ΔybdZ without iron restriction (−100 µM 2,2′-dipyridyl) is shown in black, while the growth of Para9150ΔybdZ under iron restriction (+100 µM 2,2′-dipyridyl) is shown in blue. Each point and error bar indicates the mean ± s.e.m. of OD600, where n = 3 biologically independent experiments for all curves except SUFST4/74 in red, which was derived from n = 7 biologically independent experiments.
We were intrigued by an LPS modification gene cluster containing rfbE (Fig. 2e), a typhoid-specific gene that modifies the O-antigen terminal sugar to tyvelose56,57. While tyvelose is used for serotyping, its biological function in typhoidal Salmonella remains unclear. Tn insertions in rfbE sensitize typhoidal Salmonella to bile acids, protamine and polymyxin B (Fig. 2f and Supplementary Fig. 9). To validate these results, we deleted rfbE in S. Typhi Ty2 and found that Ty2ΔrbfE exhibited decreased survival in response to bile acids (Fig. 2g) and protamine (Fig. 2h) compared with Ty2WT, suggesting that this unique tyvelose moiety protects Typhi from membrane-insulting stresses and antibiotics.
A typhoid-specific phenotype for metQ during reactive chlorine stress
metIQN encodes the main methionine transporter in Salmonella and was identified as a gene cluster with typhoid-specific fitness effects by SAFE58 (Fig. 3a,b). Tn insertions in metIQN rendered both S. Typhi Ty2 and S. Paratyphi A highly susceptible (~sevenfold) to reactive chlorine stress (RCS) but did not sensitize S. Typhimurium ST4/74 (Fig. 3c and Extended Data Fig. 2a,b). To confirm these Rb-Tn-seq results, we deleted metQ in both S. Typhi Ty2 and S. Typhimurium ST4/74 and found that Ty2ΔmetQ had a more severe growth defect under RCS than ST4/74ΔmetQ (Fig. 3d,e). Deleting metQ in six S. Typhi clinical isolates, including CT18 and multidrug resistance strains belonging to the H58 lineage (ISP-04-06979, ISP-03-07467, E03-9804, ISO(98S) and E03-4983)59, also increased RCS sensitivity (Extended Data Fig. 2c–h).
RCS kills bacteria by oxidizing cysteine (Cys) and methionine (Met)60. Salmonella replenishes these sulfated amino acids by either importing them via dedicated transporters like MetIQN or through de novo synthesis58. Given the increased susceptibility of Ty2ΔmetQ to RCS, we reasoned that endogenous Cys/Met in Typhi may be impaired. To test this hypothesis, we cultured both S. Typhimurium ST4/74 and S. Typhi Ty2 in defined minimal medium. Both strains grew when all 20 amino acids (AA) were added (Fig. 3f,g). However, in medium lacking Cys/Met (18AA −Cys −Met), ST4/74 continued to grow, while Ty2 failed to grow (Fig. 3f,g), suggesting that S. Typhi cannot synthesize Cys/Met endogenously.
To pinpoint the compromised part of the Cys/Met synthesis pathway in S. Typhi (Fig. 3h), we expressed plasmid-borne functional versions of each operon in this pathway (cysIJH, cysCND, cysK, cysPTWAM) from S. Typhimurium in S. Typhi Ty2. Only cysIJH from ST4/74 restored Ty2 growth in the 18AA medium lacking Cys/Met (Fig. 3i). Furthermore, deleting cysIJ in ST4/74 ablated growth of this isolate in medium without Cys/Met (Extended Data Fig. 2i), emphasizing the importance of CysI/J in both nontyphoidal and typhoidal Salmonella under Cys/Met limitation. Importantly, cysIJST4/74 expression rescued Ty2ΔmetQ growth under a lethal dose of RCS (Fig. 3j). In contrast, expressing cysIJ from Ty2 only partially rescued S. Typhi growth in Cys/Met-deficient media (Extended Data Fig. 2j), and weakly in Ty2ΔmetQ under lethal RCS exposure (Fig. 3j), further suggesting that CysIJTy2 function is impaired. Collectively, these findings indicate the increased sensitivity of Ty2ΔmetQ to RCS is driven by defects in endogenous Cys/Met synthesis.
A serovar-specific phenotype for ybdZ under iron restriction
We identified a paratyphoid-specific gene network displaying fitness defects, primarily consisting of iron-related genes and featuring an unannotated gene RS10805, sharing ~60% identity with ybdZ in Escherichia coli (Fig. 4a and Extended Data Fig. 3a–c), which enhances enterobactin production61. Quantitative real-time (qRT)-PCR showed an increase in ybdZ expression of ~200-fold during iron restriction in S. Paratyphi A (Fig. 4b), consistent with a role during iron limitation in Salmonella. Tn insertions in ybdZ caused a more pronounced fitness defect under iron limitation in S. Paratyphi A than in S. Typhimurium (Fig. 4c). To confirm this Rb-Tn-seq result, we deleted ybdZ in both S. Paratyphi A 9150 and S. Typhimurium ST4/74 and observed that Para9150ΔybdZ had a stronger growth defect under iron limitation than ST4/74ΔybdZ (Fig. 4d,e). Deleting ybdZ in other Paratyphi A strains (Para11511 and Para12176) also led to heightened sensitivity to iron restriction, and exogenous iron rescued these phenotypes (Extended Data Fig. 3d,e).
To understand why Para9150ΔybdZ exhibits a more severe growth defect under iron limitation than ST4/74ΔybdZ, we investigated the impact of pseudogenes in typhoidal Salmonella involved in iron acquisition, including fhuA, fhuE, sufD and sufS7. SufS/D are components of the SUF complex, one of two multiprotein complexes that synthesizes iron-sulfur clusters. In E. coli, SUF contributes to survival during iron restriction62,63,64. Strikingly, expressing functional versions of these genes from S. Typhimurium in Para9150ΔybdZ revealed that only SUFST4/74 expression restored growth during iron restriction (Fig. 4f), suggesting that functional SUF is sufficient to rescue this growth defect. To determine whether SUF is also necessary for survival under iron limitation in a ΔybdZ mutant background, we constructed a double deletion mutant of ybdZ and sufSD (ΔybdZΔsufSD) in Typhimurium ST4/74 and found that it was highly sensitive to iron restriction (Extended Data Fig. 3f), confirming that SUF contributes to survival under iron restriction in both nontyphoidal and typhoidal Salmonella. Notably, sufS and/or sufD are pseudogenes in all deposited Paratyphi A sequences on BioCyc, indicating the increased sensitivity of Paratyphi A to iron restriction is likely a general feature of Paratyphi A.
Serovar-specific fitness during macrophage infection
We aimed to identify serovar-specific phenotypes in a host-associated setting. Salmonella replication within mammalian macrophages is a key feature of this pathogen65,66,67. Despite previous Tn-seq studies examining Salmonella fitness in macrophages27,28,29, a systematic comparison of intracellular fitness profiles between generalist and human-restricted Salmonella within human macrophages is lacking. To address this gap, we performed Rb-Tn-seq on human THP-1 macrophages infected with our four barcoded libraries, using a multiplicity of infection and infection duration favoring Salmonella replication while minimizing host cell death (Fig. 5a and Supplementary Fig. 10a–g). Two macrophage passages were conducted to enrich subtle phenotypes (Fig. 5a). Consistent with published work, the absence of Vi capsule increased S. Typhi uptake into macrophages68 (Supplementary Fig. 10h); thus, we performed the Typhi Rb-Tn-seq in a ΔVi capsule background to increase intracellular bacteria.
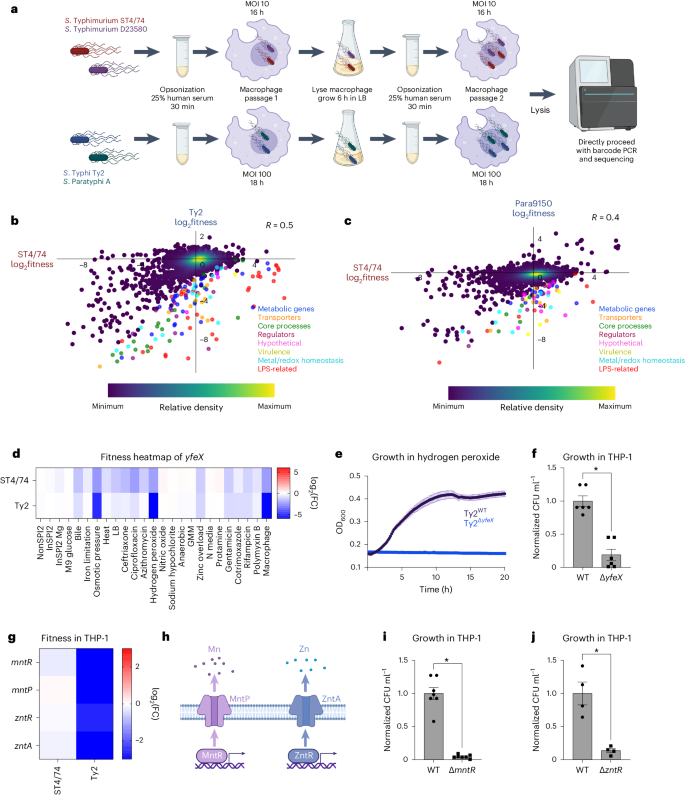
a, Schematic of macrophage infections, showing opsonization, cell infection, lysis and passaging. b,c, Pearson’s correlation (R) of gene fitness changes between the S. Typhimurium ST4/74 and S. Typhi Ty2 (b) or S. Paratyphi A 9150 (c) macrophages Rb-Tn-seq experiments, with serovar-specific changes in fitness colored according to functional class. Blue, metabolic genes; orange, transporters; green, core process genes (for example, replication, transcription, translation); burgundy, regulators; pink, hypothetical genes; gold, virulence genes; light blue, metal/redox homeostasis genes; red, LPS-related genes; dark purple, all other genes. Data are shown as a density scatterplot, where colors range from dark purple (low) to yellow (high), representing the kernel density estimation from low to high density. d, Heatmap showing the fitness values of Tn insertions in yfeX across 24 stress conditions for S. Typhimurium ST4/74 and S. Typhi Ty2. Color gradient is derived from the log2(FC) from each condition in the Rb-Tn-seq experiments. e, Growth curves of Ty2WT (black) and Ty2ΔyfeX (blue) when exposed to 250 µM hydrogen peroxide, with reads taken at OD600 once every 10 min. Each point and error bar indicates the mean ± s.e.m. of OD600, derived from n = 3 biologically independent experiments. f, Normalized intracellular Ty2WT and Ty2ΔyfeX bacterial counts recovered after 5 h of infection of LPS-activated THP-1 cells (P = 4.03 × 10−5). g, Heatmap showing the fitness values of Tn insertions in metal homeostasis genes during THP-1 infection by S. Typhimurium ST4/74 and S. Typhi Ty2. Color gradient is derived from the log2(FC) from each condition in the Rb-Tn-seq experiments. h, Schematic of Mn and Zn homeostasis systems in Salmonella. i, Normalized intracellular Ty2WT and Ty2ΔmntR bacterial counts recovered after 5 h of infection in LPS-activated THP-1 cells (P = 2.05 × 10−7). j, Normalized intracellular Ty2WT and Ty2ΔzntR bacterial counts after 5 h of infection in LPS-activated THP-1 cells (P = 0.00259). For all macrophage CFU plots (f, i and j), bars indicate the mean ± s.e.m. of the normalized bacterial count recovered from macrophages, derived from n = 6 biologically independent experiments (f), n = 7 biologically independent experiments (i) and n = 4 biologically independent experiments (j). Significance was calculated using a two-tailed t-test; *P < 0.01.
We used agglomerative clustering to generate a heatmap from these macrophage experiments, retaining all genes with significant fitness effects (P < 0.05) in at least one serovar (Supplementary Fig. 11). This heatmap revealed that Typhi and Paratyphi A cluster together while Typhimurium ST4/74 and D23580 cluster together (Supplementary Fig. 11), suggesting that the genes involved in macrophage infection exhibit greater similarity within typhoidal strains and nontyphoidal isolates, respectively. This heatmap captured several SPI-related gene clusters (Supplementary Table 7). Tn insertions in SPI-2 genes displayed decreased fitness in S. Typhimurium ST4/74, D23580 and S. Typhi Ty2 (Supplementary Table 7 and Supplementary Fig. 12a), consistent with previous findings highlighting the critical role of SPI-2 in intracellular S. Typhimurium28,29,65,68 and S. Typhi proliferation69. Intriguingly, no significant fitness defects in SPI-2 genes were observed in S. Paratyphi A 9150. This might suggest that Paratyphi A does not rely solely on SPI-2 for intracellular survival69, or could reflect the low intracellular biosynthetic capacity of S. Paratyphi A70. Mutations in the Typhimurium-specific effectors sseK1 and sseK3 led to decreased fitness in ST4/74 and D23580 within macrophages (Supplementary Data 14). Mutations in the SPI-1 encoded sitABCD Fe/Mn import system decreased Typhi and Paratyphi fitness, suggesting high sensitivity of typhoidal Salmonella to intracellular perturbations in Fe/Mn pools (Supplementary Data 14). Similarly, mutations in the SPI-3 encoded Mg2+ importers mgtB and mgtC decreased S. Paratyphi A 9150 fitness during macrophage infection (Supplementary Data 14).
Beyond SPI-genes, Tn insertions in several two-component signaling genes (for example, phoPQ, envZ/ompR, arcAB) and redox-related genes (for example, trxA, trxB, sodA, oxyR) decreased fitness during macrophage infection (Supplementary Data 14). In contrast, increased fitness was observed with Tn insertions in fepE in S. Paratyphi A 9150 (Supplementary Data 14). The regulation of very long O-antigen chains by FepE in Paratyphi71,72 indicates a potential effect of this modified LPS structure during macrophage uptake, akin to the role of Vi capsule in reducing Typhi phagocytosis68. Tn insertions in many LPS genes increased fitness in S. Typhimurium ST4/74 and D23580, consistent with previous studies highlighting enhanced invasiveness of O-antigen deficient S. Typhimurium (Supplementary Fig. 12b and Supplementary Table 7)29. Conversely, mutations in these O-antigen genes decreased Typhi fitness, emphasizing the importance of the O-antigen layer in shielding typhoidal isolates from membrane-disrupting stresses. For example, Ty2ΔrfbE, lacking its O-antigen tyvelose moiety (Fig. 2f), exhibited reduced fitness within macrophages (Supplementary Fig. 13a). Intriguingly, Tn insertions in chemotaxis genes (for example, cheARWY, tar) increased fitness in all four isolates (Supplementary Fig. 12c and Supplementary Table 7).
To identify serovar-specific fitness changes during macrophage infection, we correlated the fitness profiles between S. Typhimurium ST4/74 and the other isolates; the highest correlation was observed between the two Typhimurium isolates (R = 0.6; Supplementary Fig. 14), followed by Typhi (R = 0.5; Fig. 5b) and Paratyphi A (R = 0.4; Fig. 5c). We identified dozens of genes with typhoidal-specific changes in fitness, defined as genes that had FC greater than fourfold higher in S. Typhi Ty2 (Supplementary Table 8) or S. Paratyphi A 9150 (Supplementary Table 9) compared with S. Typhimurium ST4/74. For example, Tn insertions in yfeX, encoding a putative iron-dependent peroxidase, strongly reduced fitness within THP-1 cells for S. Typhi Ty2 and S. Paratyphi A 9150 (~120× and ~30× lower, respectively) and moderately for S. Typhimurium ST4/74 (~4×) (Fig. 5d). Accordingly, deleting this putative peroxidase in S. Typhi (Ty2ΔyfeX) increased sensitivity to 250 µM H2O2 (Fig. 5e), and impaired survival in LPS-stimulated THP-1 macrophages compared with Ty2WT (Fig. 5f and Supplementary Fig. 13b).
Tn insertions in genes involved in manganese (Mn) and zinc (Zn) homeostasis induced stronger fitness defects in S. Typhi during macrophage infection than in the other serovars (Fig. 5g–h). To confirm these results, we deleted mntR and zntR in S. Typhi Ty2 and observed that these mutants survived worse within LPS-activated THP-1 macrophages compared with the wild-type (Fig. 5i–j and Supplementary Fig. 13c,d), indicating heightened sensitivity to changes in intracellular Mn2+ and Zn2+ levels in human-restricted Salmonella.
A typhoid-specific phenotype for mntR under stress
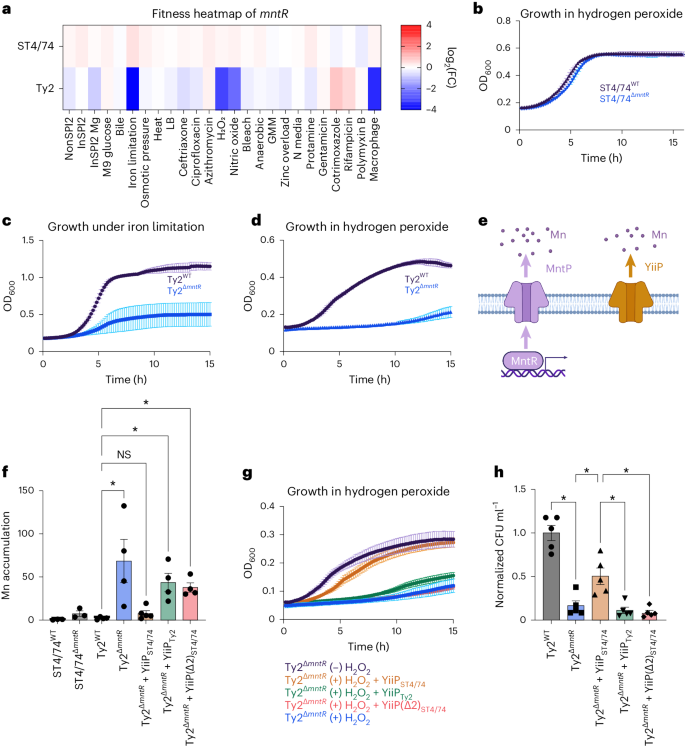
We sought to understand why S. Typhi Ty2ΔmntR has decreased intramacrophage survival. Tn insertions in mntR rendered S. Typhi sensitive to iron limitation, H2O2 and nitric oxide (NO) (Fig. 6a)—all of which all encountered within macrophages73,74,75. In contrast, Tn insertions in mntR had no impact on the fitness of other serovars (Fig. 6a and Extended Data Fig. 4a). Deleting mntR in both S. Typhimurium ST4/74 and S. Typhi Ty2 validated these results; ST4/74ΔmntR did not have a growth defect with H2O2 (Fig. 6b), whereas Ty2ΔmntR exhibited marked growth defects under iron restriction (Fig. 6c) and H2O2 (Fig. 6d), confirming its serovar-specific phenotype. Deleting mntR in six additional S. Typhi strains sensitized each strain to H2O2, indicating conservation of this phenotype across clinical Typhi isolates (Extended Data Fig. 4b–g).
a, Fitness heatmap of mntR Tn insertions for S. Typhimurium ST4/74 and S. Typhi Ty2. Color gradient is derived from the log2(FC) for each condition from Rb-Tn-seq. b, Growth of ST4/74WT (black) and ST4/74ΔmntR (blue) when exposed to 250 µM H2O2, derived from n = 4 biologically independent experiments. c, Growth of Ty2WT (black) and Ty2ΔmntR (blue) when exposed to 100 µM 2,2′-dipyridyl, derived from n = 4 biologically independent experiments. d, Growth of Ty2WT (black) and Ty2ΔmntR (blue) when exposed to 250 µM H2O2, derived from n = 3 biologically independent experiments. e, Schematic of Mn efflux systems in Salmonella. The MntR/MntP system is shown in purple, while YiiP is in orange. f, ICP-OES of Mn accumulation in WT and mutant strains of S. Typhimurium ST4/74 and S. Typhi Ty2 under Mn challenge, shown as the ratio of accumulated Mn in the (+) versus (−) Mn challenged samples, normalized to viable cell count. Each point and error bar indicates the mean ± s.e.m. of normalized Mn and is derived from n = 3 (ST4/74 strains), n = 4 (Ty2WT, Ty2ΔmntR, Ty2ΔmntR+YiiPTy2, Ty2ΔmntR+YiiP(Δ2)ST4/74) and n = 5 (Ty2ΔmntR+YiiPST4/74) biologically independent experiments. g, Growth curves of Ty2ΔmntR and complementation strains of Ty2ΔmntR under 250 µM H2O2. For controls, Ty2ΔmntR grown without H2O2 is in black, while Ty2ΔmntR growth with H2O2 is in blue, both derived from n = 3 biologically independent replicates. For complementation curves, YiiP(Δ2)ST4/74 is in red (n = 5 biologically independent replicates), while YiiPST4/74 is in orange and YiiPTy2 is in green, both derived from n = 7 biologically independent replicates. h, Normalized intracellular bacterial counts recovered from LPS-activated macrophages, with Ty2WT in gray and Ty2ΔmntR in blue. For complementation strains, YiiPST4/74 is in orange, YiiPTy2 is in green and YiiP(Δ2)ST4/74 is in red. Bars indicate the mean ± s.e.m. of the normalized bacterial count recovered from macrophages with individual values shown, derived from n = 5 biologically independent experiments. For all growth curves (b–d, g), each point and error bar indicates the mean ± s.e.m. of OD600, with reads taken at OD600 once every 10 min. For f and h, significance was calculated using a one-way ANOVA; *P < 0.05, with multiple comparisons corrected by the Benjamini, Krieger and Yekutieli method (Supplementary Table 10).
We next explored why Ty2ΔmntR is sensitive to infection-related stress. MntR—a Mn-responsive transcription factor—activates MntP, a Mn efflux pump, to restore homeostasis76,77. Nontyphoidal and typhoidal Salmonella also encode YiiP—a constitutively active Mn pump78,79 (Fig. 6e). While MntP sequences are identical between S. Typhimurium and S. Typhi, Ty2 YiiP has a two amino acid deletion (ΔL95–F96) found in all Typhi isolates on BioCyc (Extended Data Fig. 4h), suggesting that YiiPTy2 may not be fully functional. Accordingly, we challenged WT and ΔmntR mutants of S. Typhimurium ST4/74 and S. Typhi Ty2 with 200 µM manganese and quantified intracellular Mn accumulation. Neither WT ST4/74 nor WT Ty2 accumulated Mn, indicating functional Mn efflux (Fig. 6f). The ST4/74ΔmntR modestly accumulated Mn (around fourfold), suggesting that YiiP removes most intracellular Mn even without MntR in this isolate (Fig. 6f). In contrast, Ty2ΔmntR accumulated high Mn (~70-fold), indicating severely impaired Mn efflux without MntR, likely due to a nonfunctional YiiP efflux pump (Fig. 6f).
To investigate whether mutated YiiP underlies the fitness defects of Ty2ΔmntR, we expressed functional YiiP from S. Typhimurium in Ty2ΔmntR; this complemented strain no longer accumulated Mn during Mn challenge (Fig. 6f) or exhibited strong sensitivity to H2O2 (Fig. 6g). In contrast, expressing YiiP from Ty2 in the Ty2ΔmntR strain still resulted in high Mn accumulation during Mn challenge (Fig. 6f) and sustained sensitivity to H2O2 (Fig. 6g), further indicating that YiiP in S. Typhi is likely nonfunctional. Similarly, introducing the mutated YiiPST4/74 pump (YiiP(Δ2)ST4/74), with an in-frame deletion of L95–F96, into Ty2ΔmntR failed to correct growth defects under Mn or H2O2 stress (Fig. 6f–g). Moreover, deleting either full-length yiiP or just the L95–F96 sequence of this pump in ST4/74ΔmntR sensitized ST4/74 to both Mn and H2O2 stress (Extended Data Fig. 4i–j), indicating that YiiP is both necessary and sufficient for Salmonella survival to macrophage-associated stresses. Importantly, expressing YiiPST4/74 rescued Ty2ΔmntR survival in LPS-activated THP-1 macrophages to a significantly higher extent than expressing yiiPTy2 or mutated yiiP(Δ2)ST4/74 (Fig. 6h and Extended Data Fig. 4k). Collectively, our results indicate that yiiP pseudogenization in human-adapted Salmonella leads to a vulnerability during macrophage infection.